
I looked into using the cytoscape.js-cola layout with the following options: layout: ).
This discrete layout creates good results for most graphs and it supports compound nodes. I haven't found a layout that fully supports positioning nodes within the compound. The Klay layout algorithm for Cytoscape.js.
#Cytoscape klay manual
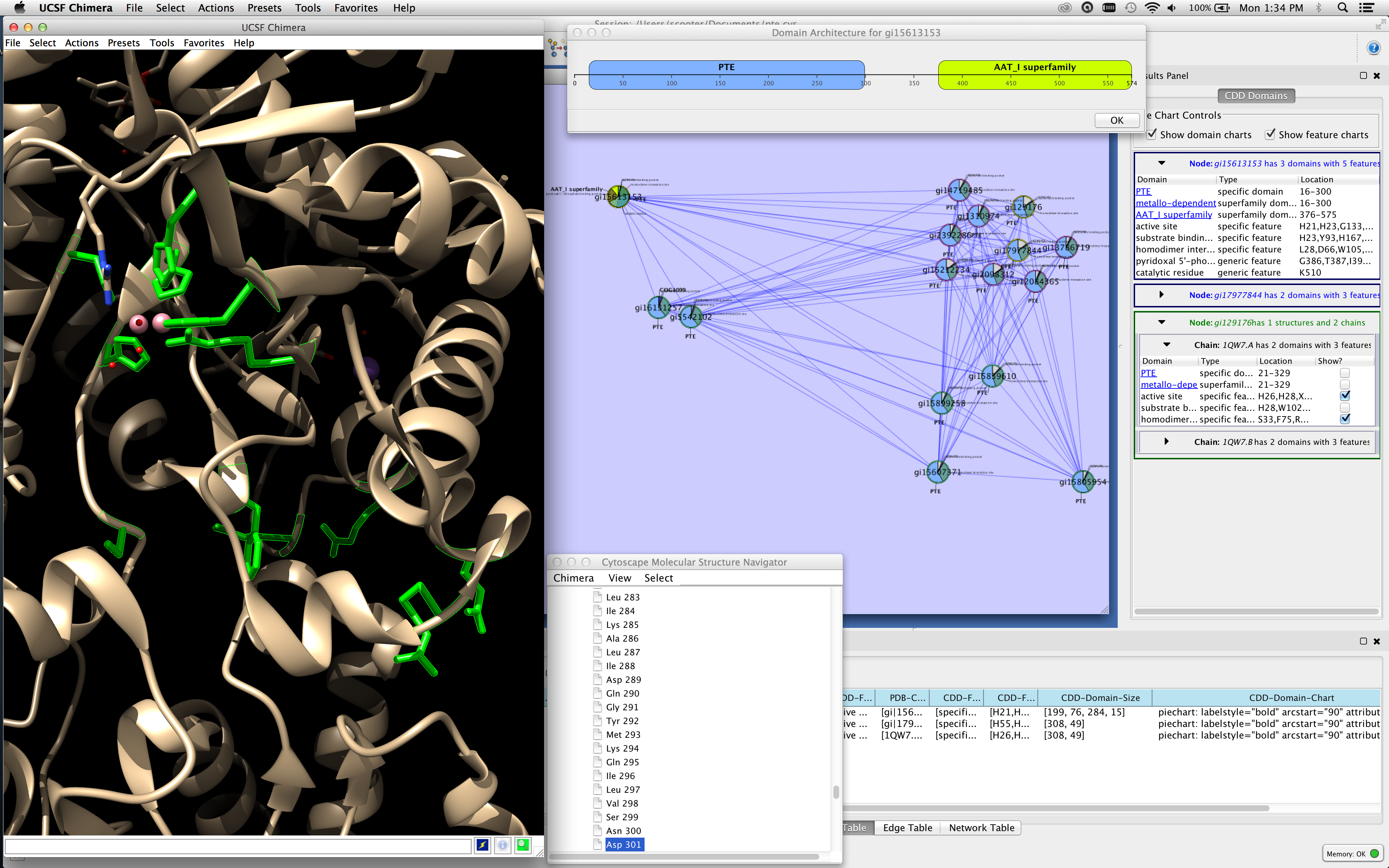
I hope it explains what I would like to end up with.įirst, I already saw this question, but as far as I understood it's for manual positioning, rather than layouts. I then specified cluster subgraphs containing all recursive components. Other elements have no rank and are hence freely positioned. Categories: automation, data integration, data visualization, local data import, pathway database, visualization. In dot I tried to add some form of flow within the system ( rankdir=LR ) by positioning the inputs on the left ( rank=source ) and the outputs on the right ( rank=sink ). A Cytoscape app to connect to a Neo4j database and execute extensions of the Neo4j database. In my graph I have compound nodes that represent components.Ĭomponents contain other components and/or states, transitions, variables.Įach component can specify inputs and outputs. The Klay layout algorithm for Cytoscape.
#Cytoscape klay install
You can install using 'npm i cytoscape-klay' or download it from GitHub, npm.
#Cytoscape klay license
cytoscape.js-klay has no bugs, it has no vulnerabilities, it has a Permissive License and it has low support. However, I am struggling to find a layout/config that supports what dot calls rank. cytoscape.js-klay is a JavaScript library. When a graph is first loaded, the user should be presented with a default layout though. For example, the first frame might be of a network zoomed out and the second frame might focus on a specific group of nodes (see Figure 1). Download the library: via npm: npm install cytoscape-klay, via bower: bower install cytoscape-klay, or. Cytoscape.js 3.2.0 Klay 0.4.1 Usage instructions. The tool allows you to take a series of snapshots (CyAnimator calls them frame s) of Cytoscape networks. The Klay layout algorithm for Cytoscape.js.

This project is set up to automatically be published to npm and bower.I am trying to use cytoscape to replace my dot output and make it interactive (move nodes and compounds, expand/collapse compounds, etc.) CyAnimator is a Cytoscape app that provides a tool for simple animations of Cytoscape networks. There are two major classes of layouts: (1) Force-directed (e.g. all builds use babel, so modern ES features can be used in the src. The Klay library does not have facilities to set restrictions on node positions. npm run lint : Run eslint on the source.npm run dev : Automatically build on changes with live reloading with webpack dev server.you must already have an HTTP server running) This tutorial presents a scenario of how expression and network data can be combined to tell a biological story and includes these concepts: Visualizing networks using expression data.
#Cytoscape klay software
You may want to use klay for its smaller file size, if the layered algorithm of elk does not differ significantly for your dataset. Cytoscape is an open source software platform for integrating, visualizing, and analyzing measurement data in the context of networks. It is the predecessor to the layered algorithm in elk. npm run watch : Automatically build on changes with live reloading (N.b. klay: The klay layout is a traditional hierarchical layout.NodeDimensionsIncludeLabels : false, // Boolean which changes whether label dimensions are included when calculating node dimensionsĪnimate : false, // Whether to transition the node positions animateFilter : function( node, i ), // Edges with a non-nil value are skipped when geedy edge cycle breaking is enabledĬy.


 0 kommentar(er)
0 kommentar(er)
